The past few years have witnessed the discovery of both very large and small genomes in different groups of organisms. Here are some highlights from this research.
The first represents the largest genome so far reported for a crustacean, in the Arctic-dwelling amphipod Ampelisca macrocephala. The genome of this small invertebrate is a whopping 63.2 billion base pairs, or about 20 times larger than the human genome (Rees et al. 2007). Again, this sort of observation should dispel the notion that all non-coding DNA is functional for protecting against mutagens or some such thing.
The second interesting finding is of the largest viral genome so far discovered. The virus, dubbed Mimivirus, was sufficiently odd that it was originally assumed to be a bacterium when first observed, but on closer examination was found to be a virus. Its genome size is estimated as 1.2 million bases, which is larger than the genome of many bacteria (Raoult et al 2007). So, now there is overlap in reported genome sizes between viruses and bacteria, which goes along with the known overlap between the genome sizes of bacteria and eukaryotes (Gregory 2005).
And now for some small genomes. More specifically, the smallest flowering plant genome, that of Genlisea margaretae at a mere 63 million base pairs, less than half the size of the previous record holder, Arabidopsis thaliana at about 157 million base pairs. This increases the range in angiosperm genome sizes to more than 2,000-fold. (In animals the total range is about 3,300-fold; Gregory et al. 2007).
The smallest insect genome so far estimated was reported fairly recently as well. It belongs to Caenocholax fenyesi, a twisted-wing parasite, and is a mere 108 million base pairs (Johnston et al. 2004). Not to spoil the fun, but my lab has also found genome sizes this small in other groups, though these have not yet been published. The largest insect genome size known is found in the mountain grasshopper Podisma pedestris at 16.6 billion base pairs (Westerman et al. 1987).
The smallest eukaryotic genome known to date is that of the protist Encephalitozoon intestinalis, a parasitic microsporidian with a genome size of only 2.3 million base pairs, which is smaller than that of many bacteria (Vivarès and Méténier 2000). The smallest free-living eukaryote genome size is found in Ostreococcus tauri at 12.6 million base pairs (Derelle et al. 2006). The largest reliable protozoan genome size estimate reported to date is 97.8 billion base pairs in the dinoflagellate Gonyaulax polyedra (Shuter et al. 1983). That is a more than 33,000-fold range among protists.
It should be pointed out that the largest published eukaryote genome size estimate is 1,400 billion base pairs (400 times larger than human) in the free-living amoeba Chaos chaos (Friz 1968), although the largest genome size is often attributed to Amoeba dubia at 700 billion base pairs based on the same study. These data are not generally considered reliable, for several reasons. First, these values for amoebae were based on rough biochemical measurements of total cellular DNA content, which probably includes a significant fraction of mitochondrial DNA. Second, Friz’s (1968) value of 300pg for Amoeba proteus is an order of magnitude higher than those reported in subsequent studies (Byers 1986). Third, some amoebae (e.g., A. proteus) contain 500-1000 small chromosomes and are quite possibly highly polyploid (Byers 1986), in which case these values would be inappropriate for a comparison of haploid genome sizes among eukaryotes.
Finally, the smallest genome so far known for any cellular organism also was discovered recently — that of the endosymbiotic bacterium Carsonella ruddii at a miniscule 159,662 base pairs (Nakabachi et al. 2006). This species resides within specialized cells inside the body of psyllid insect hosts. The genome is so small, and the insect and bacterium so mutually dependent, that this species blurs the lines between bacteria and organelles, and probably is similar in some ways to an intermediate stage in the evolution of other obligate intracellular symbionts turned organelles like mitochondria and chloroplasts.
The old assumption, still often repeated, that viruses have smaller genomes than bacteria which have smaller genomes than single-celled eukaryotes which have smaller genomes than multicellular eukaryotes is beginning to wear thin. The pattern remains in a general sense, but focusing only on such a coarse scale overlooks a significant amount of diversity within, and increasingly apparent overlap between, groups of life.
___________
Readers interested in exploring genome size data can check out the various online databases for more.
References
Byers, T.J. 1986. Molecular biology of DNA in Acanthamoeba, Amoeba, Entamoeba, and Naegleria. International Review of Cytology 99: 311-341.
Derelle, E., C. Ferraz, S. Rombauts, P. Rouzé, A.Z. Worden, S. Robbens, F. Partensky, S. Degroeve, S. Echeynié, R. Cooke, Y. Saeys, J. Wuyts, K. Jabbari, C. Bowler, O. Panaud, B. Piégu, S.G. Ball, J.P. Ral, F.Y. Bouget, G. Piganeau, B. De Baets, A. Picard, M. Delseny, J. Demaille, Y. Van de Peer, H. Moreau. 2006. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proceedings of the National Academy of Sciences of the USA 103: 11647-11652.
Friz, C.T. 1968. The biochemical composition of the free-living amoebae Chaos chaos, Amoeba dubia, and Amoeba proteus. Comparative Biochemistry and Physiology 26: 81-90.
Gregory, T.R. 2005. Synergy between sequence and size in large-scale genomics. Nature Reviews Genetics 6: 699-708.
Gregory, T.R., J.A. Nicol, H. Tamm, B. Kullman, K. Kullman, I.J. Leitch, B.G. Murray, D.F. Kapraun, J. Greilhuber, and M.D. Bennett. 2007. Eukaryotic genome size databases. Nucleic Acids Research 35 (Suppl. 1): D332-D338.
Greilhuber, J., T. Borsch, K. Müller, A. Worberg, S. Porembski, W. Barthlott. 2006. Smallest angiosperm genomes found in lentibulariaceae, with chromosomes of bacterial size. Plant Biology 8: 770-777.
Johnston, J.S., L.D. Ross, L. Beani, D.P. Hughes, and J. Kathirithamby. Tiny genomes and endoreduplication in Strepsiptera. Insect Molecular Biology 13: 851-585.
Nakabachi A, A. Yamashita, H. Toh, H. Ishikawa, H.E. Dunbar, N.A. Moran, and M. Hattori. 2006. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314: 267.
Raoult, D., B. La Scola, and R. Birtles. 2007. The discovery and characterization of Mimivirus, the largest known virus and putative pneumonia agent. Clinical Infectious Diseases 45: 95-102.
Rees, D.J., F. Dufresne, H. Glémet, and C. Belzile. 2007. Amphipod genome sizes: first estimates for Arctic species reveal genomic giants. Genome 50: 151-158.
Vivarès, C.P. and G. Méténier 2000. Towards the minimal eukaryotic parasitic genome. Current Opinion in Microbiology 3: 463–467.
Westerman, M., N.H. Barton, and G.M. Hewitt (1987). Differences in DNA content between two chromosomal races of the grasshopper Podisma pedestris. Heredity 58: 221-228
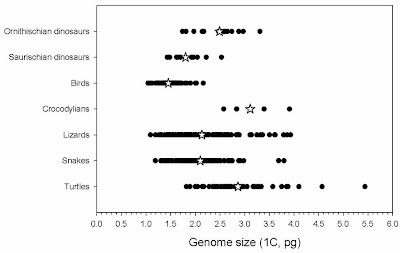 The most reasonable interpretation of this is that genomes began to shrink in saurischian dinosaurs, possibly in association with endothermy, and then they shrank more along with the evolution of powered flight.
The most reasonable interpretation of this is that genomes began to shrink in saurischian dinosaurs, possibly in association with endothermy, and then they shrank more along with the evolution of powered flight.