There sometimes is confusion surrounding the units employed in genome size publications. Genome sizes — the amount of DNA per copy of a genome — have traditionally been given in units of mass, namely picograms (1 pg = 10-12 g). More recently, people have been interested in knowing the number of base pairs per genome rather than genomic mass, which makes sense if one wishes to sequences those base pairs.
The fact is that essentially all genome size measurements represent relative estimates based on the density of stain or fluorescence of dye as compared to a standard (more on this in a later post), with the exception of truly complete genomic sequencing, which is very uncommon for eukaryotes. The units into which these relative data are converted is simply a matter of preference, and if one knows the mass of a given nucleotide then one can easily convert between picograms and base pairs. Indeed, Dolezel et al. (2003) did this calculation based on the following data:
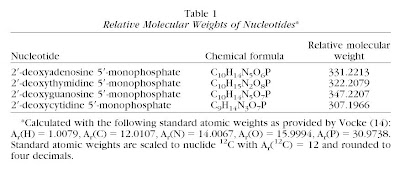
The net result is that one easily can convert between picograms and base pairs as follows:
Yes, there will be a little bit of error involved if there are biases toward AT or GC in the genome. However, “by using the data in Table 1, relative weights of nucleotide pairs can be calculated as follows: AT = 615.3830 and GC = 616.3711, bearing in mind that one phosphodiester linkage involves a loss of one H2O molecule” (Dolezel et al. 2003). In other words, the difference is very slight and is negligible relative to the experimental error inherent in any genome size estimate.
To put it very simply, the units are interchangeable with
1 pg = 978 Mbp.
————–
Dolezel, J., J. Bartos, H. Voglmayr, and J. Greilhuber. 2003. Nuclear DNA content and genome size of trout and human. Cytometry 51A: 127-128.
