The title “Doctor” and the abbreviated prefix “Dr.” come from the Latin for “teacher”, and are traditionally bestowed on those who have earned a doctoral degree, the highest academic degree attainable. The suffix Ph.D. is an abbreviation for Philosophiæ Doctor (L. “Teacher of philosophy”), with “philosophy” from the Greek for “love or pursuit of wisdom”. The Ph.D. is awarded in most academic disciplines, including science (in many cases, the D.Sc., or Doctor of Science, is awarded as an honour for special accomplishment). Medical professionals may also hold the title “Doctor” even though they may do little or no teaching, with common suffixes being M.D. (Medicinae Doctor, or Doctor of Medicine), D.V.M. (Doctor of Veterinary Medicine), D.D.S. (Doctor of Dental Surgery), and so on. It is also possible to obtain the title “Doctor” in areas such as homeopathic medicine or chiropractic. In other words, the title alone does not provide much information about what the individual’s qualifications are. However, one can reasonably argue that the prestige attached to it comes from the fact that individuals such as medical doctors and scientists who hold the title are typically ranked among those with greatest prestige.
For the past several years, i.e., since completing my Ph.D. degree, my official title has been “Dr.” and not “Mr.”. Some people become upset if you call them “Mr.” (or “Ms.”) instead of “Dr.”, but I generally try not to make too much of this. In academic settings, I think a good rule is that people who do not hold the degree, or colleagues in formal settings, should address those with the degree by their official title of “Dr.”. On the other hand, in informal settings with colleagues who hold the same title, or with students with whom one is familiar (e.g., graduate students in one’s own group), using the title simply becomes awkward and first names are appropriate. I will admit that it did take me some time to get used to calling my advisors by their first names, and I have since observed this in several of my students. Outside of academic settings, I generally don’t correct people if they call me “Mr.”, and I don’t introduce myself with the title. I believe most of my colleagues take a similar approach, though of course there is some variation.
Two recent discussions on science blogs have prompted this post, in case you were curious as to why I was bothering to tell you this. Both relate to what I am calling the “Dr. Credibility principle”, which is an attempt to gain undeserved credibility simply through invoking the title.
The first is the example of Dr. Sharon Moalem, author of the book Survival of the Sickest. It appears Dr. Moalem has been misrepresenting his credentials, making it seem as though he has obtained a medical degree, when in fact he has not (though he his a med student). PZ Myers has already discussed the book, which seems to be largely based on non-scientific and pseudoscientific arguments. I had a feeling that this would be so right away. Why? Because he used the title “Dr.” on the cover.
You see, scientists almost never use the prefix “Dr.” on their book covers. It doesn’t matter if it is a popular book or a technical one. If you have science books, go see for yourself. We just don’t do it. In fact, I am willing to extend the “Dr. Credibility principle” to claim that whenever you see someone making a big deal out of their title, especially on a book cover, you can usually bet it is because the content of the book cannot stand on its own without a deflecting appeal to authority. Yet, as noted, those with real authority, and because of whom the title has some prestige, do not use it in that way.
The second case is the Discovery Institute’s public relations gimmick of having “scientists” sign the following “Dissent from Darwinism” statement:
We are skeptical of claims for the ability of random mutation and natural selection to account for the complexity of life. Careful examination of the evidence for Darwinian theory should be encouraged.
As I tell my students, this is so benign that I could easily sign it, and I suspect any biologist could. I too am skeptical of claims that random mutation and natural selection account for everything. And careful examination of the evidence should be encouraged in all sciences, always. Nevertheless, as John Lynch has demonstrated, people with qualifications in evolutionary biology don’t sign this list because it is obviously intended to confuse the uninitiated.
Compare this with a statement that many biologists (though only those with the name Steve or its derivatives) are only too happy to put their names on:
Evolution is a vital, well-supported, unifying principle of the biological sciences, and the scientific evidence is overwhelmingly in favor of the idea that all living things share a common ancestry. Although there are legitimate debates about the patterns and processes of evolution, there is no serious scientific doubt that evolution occurred or that natural selection is a major mechanism in its occurrence. It is scientifically inappropriate and pedagogically irresponsible for creationist pseudoscience, including but not limited to “intelligent design,” to be introduced into the science curricula of our nation’s public schools.
The Discovery Institute has managed to find only 700 signatories since 2001. The NCSE’s list, which is meant only as a parody, has 860 Steves.
Most of the individuals who have signed the Discovery Institute’s list have degrees in engineering (see the Salem conjecture), chemistry, physics, or computer science, though several at least have biological training, albeit in physiology or molecular biology. Some of them are not scientists of any sort. The only unifying feature appears to be that they hold a Ph.D. in something. Once again, it’s the “Dr. Credibility principle”: If one has the title, one must be an authority on the issue under discussion.
The Dr. Credibility principle finds it most extraordinary application in the Orwellian-entitled Physicians and Surgeons for Scientific Integrity. Who can join, you ask? “Any person with an M.D., D. O., D.D.S., D.M.D., D.V.M. or equivalent degree may become a physician/surgeon member of PSSI”. Personally, I don’t think anyone should take a medical doctor, dentist, or veterinarian as an authority on biology any more than they should let biologists like me prescribe drugs or perform a root canal. The only possible explanation is that this is another misguided appeal to authority. Unfortunately, many people are liable to buy into it, especially in groups like anti-evolutionists, where authority seems to trump data regularly.
Let me say it clearly. Having the title “Dr.” does not make anyone an authority on anything except, one would hope, the field in which they obtained a degree. Anything else is just playing doctor to gain undue credibility.
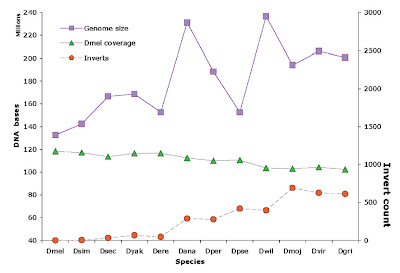 Figure 1. Drosophila species assemblies, showing assembly sizes and coverage of these by D. melanogaster genome DNA (top and middle lines, in megabases, left ordinate), and counts of chromosome segments inverted relative to Dmel (bottom line, right ordinate). Species on abscissa are taxonomically ordered with Dgri most distant from Dmel.
Figure 1. Drosophila species assemblies, showing assembly sizes and coverage of these by D. melanogaster genome DNA (top and middle lines, in megabases, left ordinate), and counts of chromosome segments inverted relative to Dmel (bottom line, right ordinate). Species on abscissa are taxonomically ordered with Dgri most distant from Dmel.