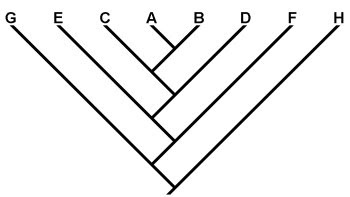
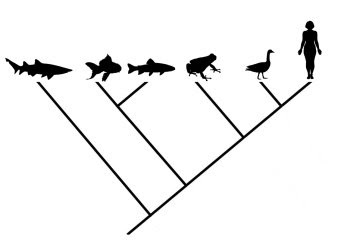
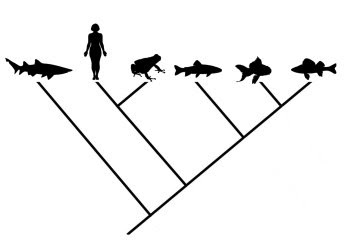
Evolutionary trees, or “phylogenies”, are a major part of modern evolutionary science. They depict hypotheses regarding the relationships among taxa, and are therefore important in reconstructions of the historical path of evolution (Gregory 2008a,b). Various approaches can be taken to formulating phylogenetic hypotheses, including analyses based on morphological, fossil, and/or molecular data. These methods often agree well, but sometimes one or another can throw up some surprises and challenge previous hypotheses about the relationships among groups of organisms. Reconstructing the tree of life is a difficult and complicated process, and one should expect there to be significant refinements and revisions along the way. This is especially true of the deepest branches of the tree, which are often the most difficult to resolve.
Case in point, the Tree of Life Web Project gives the following summary of deep branches among major animal lineages:
 Two things are apparent from this phylogeny. First, most of the branchings (“nodes”) are unresolved at the deepest point. This is known as a “soft polytomy”, meaning that these lineages probably did not emerge all at the same time from a distant ancestor, but rather that the data simply are not sufficient at present to determine the order of their branching. Second, sponges (Phylum Porifera) are typically considered to represent descendants of the earliest branching lineage.
Two things are apparent from this phylogeny. First, most of the branchings (“nodes”) are unresolved at the deepest point. This is known as a “soft polytomy”, meaning that these lineages probably did not emerge all at the same time from a distant ancestor, but rather that the data simply are not sufficient at present to determine the order of their branching. Second, sponges (Phylum Porifera) are typically considered to represent descendants of the earliest branching lineage.
Sponges are simple, sessile, filter-feeding animals comprised of only a handful of cell types. It has commonly been suggested that they retain characteristics similar to those of some of the earliest animals to appear. In other words, even though they are modern species, it is thought that their simplified morphology reflects the ancestral condition, having undergone little change over hundreds of millions of years of evolution.
Recently, a study by Dunn et al. (2008) using a comparatively large genetic dataset suggested something of a shake-up of the evolutionary tree at the deepest branches. Specifically, their data suggested that comb jellies (Phylum Ctenophora), rather than sponges, are modern representatives of the earliest branching lineage.
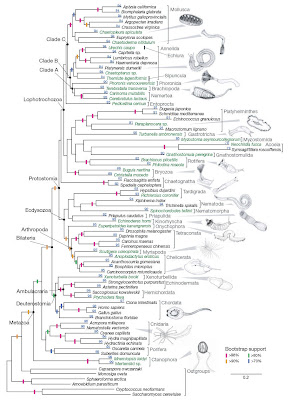 From Dunn et al. (2008). Click for larger view.
From Dunn et al. (2008). Click for larger view.Ctenophores are superficially similar to jellyfish, and are much more complex than sponges. On the face of it, finding that ctenophores are descended from the earliest-branching animals is surprising. However, this should not be taken too far, as it has in some of the press surrounding the article. For example, LiveScience presented a report entitled “Shock: First animal on Earth was surprisingly complex“. As they describe the situation,
Earth’s first animal was the ocean-drifting comb jelly, not the simple sponge, according to a new find that has shocked scientists who didn’t imagine the earliest critter could be so complex.
This interpretation illustrates a common misconception about evolutionary trees, one that I addressed in a recent paper on the topic (Gregory 2008b). Specifically, it draws the false conclusion that a modern member of an early branching lineage is very similar to the distant ancestor that it shares with other lineages. In actuality, the species under consideration are all modern species whose lineages have been evolving for exactly the same amount of time since their divergence from a common ancestor. The comb jelly lineage may have branched first, but the common ancestor from which it and the other animals lineages diverged probably looked nothing like a comb jelly. It is entirely possible that comb jellies are highly derived (i.e., very different from their early ancestor), just as other animal lineages are.
Consider the phylogeny below, which presents relationships among the living groups of echinoderms:
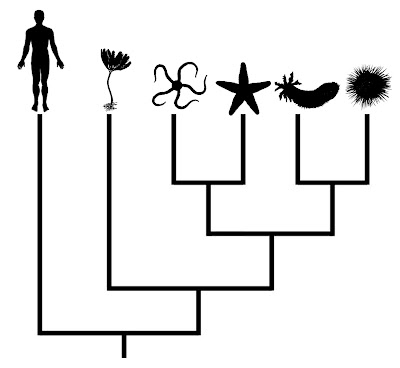 From Gregory (2008b).
From Gregory (2008b).In this case, humans are presented as the outgroup of the tree. Even though humans are shown as having branched earliest in this phylogeny, one cannot conclude that they look anything like the common ancestor shared by vertebrates and echinoderms. Unfortunately, this sort of misinterpretation is not uncommon for less obvious comparisons, even in the primary literature (Crisp and Cook 2005).
Dunn et al. (2008) provide a very reasonable interpretation of the data that does not mesh with that presented in the opening of the LiveScience report:
The placement of ctenophores (comb jellies) as the sister group to all other sampled metazoans is strongly supported in all our analyses. This result, which has not been postulated before, should be viewed as provisional until more data are considered from placozoans and additional sponges. If corroborated by further analyses, it would have major implications for early animal evolution, indicating either that sponges have been greatly simplified or that the complex morphology of ctenophores has arisen independently from that of other metazoans.
This is to say, one cannot take the finding as well established without further data, and even if it is corroborated it simply means that either ctenophores evolved their level of complexity independently of other animal lineages (i.e., the common ancestor was simple), or sponges are secondarily simplified (i.e., the common ancestor was fairly complex). Neither scenario is especially shocking, given that the sponges and comb jellies being studied are modern groups.
Strangely enough, the LiveScience story includes this information, but the implications of it apparently were not grasped:
Dunn says that two evolutionary scenarios can explain why the comb jellies would actually have been first among animals. The first is that the comb jelly evolved its complexity independent of other animals after branching off to forge its own path.
The second is that the sponge evolved its simpler form from the more complex form. This second possibility underscores the fact that “evolution is not necessarily just a march towards increased complexity,” Dunn said.
The first animals probably were not very similar to comb jellies as we now know them, namely as representatives of a lineage that has been evolving for hundreds of millions of years since diverging from those of other major animal groups. Sponges, too, have been evolving for hundreds of millions of years, and there is no reason to assume that they have not undergone any significant morphological changes in that time. Molecular data provide insights into the order of branching, but fossils are required in order to reach conclusions about morphological change or lack thereof over evolutionary timescales.
In short, one cannot assume that a modern representative of an early branching lineage is the same as the ancestor from which it is descended.
_______
Crisp, M.D. and L.G. Cook. 2005. Do early branching lineages signify ancestral traits? Trends in Ecology and Evolution 20: 122-128.
Dunn, C.W., A. Hejnol, D.Q. Matus, K. Pang, W.E. Brown, S.A. Smith, E. Seaver, G.W. Rouse, M. Obst, G.D. Edgecombe, M.V. Sorensen, S.H.D. Haddock, A. Schmidt-Rhaesa, A. Okusu, R.M. Kristensen, W.C. Wheeler, M.Q. Martindale, and G. Giribet. 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452: 745-749.
Gregory, T.R. 2008a. Evolution as fact, theory, and path. Evolution: Education and Outreach 1: 46-52.
Gregory, T.R. 2008b. Understanding evolutionary trees. Evolution: Education and Outreach 1: 121-137.
______
Update:
See also Stranger Fruit and Nimravid. I should also have noted that this comparison only includes representatives of lineages that survived to the present. Ctenophores may be members of the earliest branching living lineage (I remain skeptical, mind you), but many lineages may have diverged earlier but subsequently vanished — again, this would mean that modern ctenophores were not the first animals (and, besides, it’s quite possible that the Ediacarans were the first animals but neither sponge nor ctenophore). And the whole notion of “diverged first” is somewhat misleading — there was a split and two lineages diverged from each other; one is not a side branch of a main line.