The Royal Ontario Museum will be opening the Darwin: The Evolution Revolution exhibit today, running through August 4. The exhibit originated at the American Museum of Natural History under the curation of Niles Eldredge, and is currently touring museums in Boston, Chicago, Toronto, and London. Obviously, this would be well worth the trip.
Evolution: Education and Outreach, issue 2.
 The second issue of Evolution: Education and Outreach is available online and the various papers are free to download. The actual hard copy of the issue will be out sometime next month.
The second issue of Evolution: Education and Outreach is available online and the various papers are free to download. The actual hard copy of the issue will be out sometime next month.
My contribution is called “Understanding evolutionary trees“, which I hope will clarify some misunderstandings of this important topic.
Bird genomes.
I have been hesitant to talk about the association between bird genome size and flight, even though it has been fluttering around in the blogosphere for some time now (e.g., here and here and here). This may seem counterintuitive, since of all the bloggers interested in the issue, I have actually published articles on the subject. The reason is that a colleague and I have ostensibly been working on a paper about this subject (though it has been on the back burner for a long time) and because I have a student doing research on this issue and I did not feel it appropriate to discuss his unpublished study. But today there is another blog discussion (here and here) about how some dinosaurs already had small genomes and therefore that genome reduction was not part of the evolution of flight in the avian descendants of those dinosaurs. I figure one small clarification is useful.
Modern birds have smaller genomes than the dinosaurs are estimated to have had, strong flyers have the smallest, and flightless birds the largest.
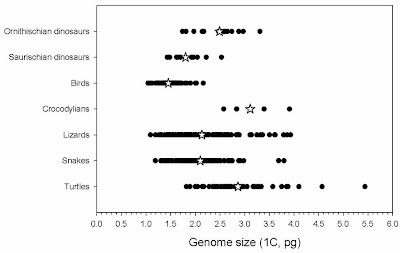 The most reasonable interpretation of this is that genomes began to shrink in saurischian dinosaurs, possibly in association with endothermy, and then they shrank more along with the evolution of powered flight.
The most reasonable interpretation of this is that genomes began to shrink in saurischian dinosaurs, possibly in association with endothermy, and then they shrank more along with the evolution of powered flight.
____________
References
Gregory, T.R. 2002. A bird’s-eye view of the C-value enigma: genome size, cell size, and metabolic rate in the class Aves. Evolution 56: 121-130.
Gregory, T.R. 2005. Genome size evolution in animals. In: The Evolution of the Genome (ed. T.R. Gregory), pp. 3-87. Elsevier, San Diego.
Organ, C.L., A.M. Shedlock, A. Meade, M. Pagel, and S.V. Edwards. 2007. Origin of avian genome size and structure in non-avian dinosaurs. Nature 446: 180-184.
Zimmer, C. 2007. Jurassic genome. Science 315: 1358-1359.
"Basic research is the lifeline of practical advances".
Unfortunate though it is, we in the scientific community seem to have to justify regularly the “relevance” of basic, or curiosity-driven, research. Case in point, the February 2008 issue of the CAUT Bulletin has a commentary by Vern Paetkau who makes the point once again that basic research underpins most future advances in applied science. I will let Dr. Paetkau speak for himself by simply referring you to his piece, which can be found here. I will, however, re-quote what I think is a superb point by Nobel laureate Arthur Kornberg:
“No matter how counter-intuitive it may seem, basic research is the lifeline of practical advances in medicine and pioneering inventions are the source of industrial strength.”
Scientists know this, but those with control of the funds often do not see it this way — and that’s a big problem for everyone, not just researchers.
Nature on the plight of Canadian science.
I love Canada. Yes, I know, we’re all human beings and national boundaries are mostly arbitrary, but I am proud of my country and its approach to its citizens and the world. I have lived in the United States and I have lived in England and I appreciated many things about both, but I am glad to be home again. I can think of no other place I would rather live.
I also love science. There are easier jobs, and higher paying ones, but there is nothing better in my mind than spending time pursuing whatever interests me, learning about how the world is and why it is that way, and sharing that knowledge — some age-old, some brand new, and some otherwise known only by me as its discoverer — with others.
The Canadian research system differs from those of some other nations in that it has always aimed to support as broad a range of scientists as possible. We traditionally did not have multi-million-dollar grants given to a select few, but rather much smaller grants distributed to a large percentage of researchers. It used to be that a good researcher — rather than a particular study — would be funded and could more or less pursue his or her interests, or could switch focus if some new and exciting topic emerged. Dollar for dollar, Canadian researchers have been incredibly productive, generating a competitive amount of knowledge with what must seem to colleagues in some other countries as pocket change.
Thus, you can imagine my dismay when I see what our current administration is doing to science in Canada. The conservative government, as noted in my previous post, have decided that we should have an elitist system in which they, rather than the scientific community, determine what a short list of priority subjects are. Not surprisingly, it’s all applied science focused in an Orwellian way on the environment and health. Only those who understand nothing about science could possibly think that the best way to find solutions to applied problems is to undermine basic research. Not to mention the reputation that this administration has when it comes to issues like environmental protection and socialized health care.

ScienceNow reported recently on some asinine new policies of the Conservatives toward research, but even before this latest development Nature pointed out the suffering of Canadian science under this government:
Science in retreat
Canada has been scientifically healthy.
Not so its government.Comparisons of nations’ scientific outputs over the years have shown that Canada’s researchers have plenty to be proud of, consistently maintaining their country’s position among the world’s top ten. Alas, their government’s track record is dismal by comparison.
When the Canadian government announced earlier this year that it was closing the office of the national science adviser, few in the country’s science community were surprised. Science has long faced an uphill battle for recognition in Canada, but the slope became steeper when the Conservative government was elected in 2006.
…
Concerns can only be enhanced by the government’s manifest disregard for science. Since prime minister Stephen Harper came to power, his government has been sceptical of the science on climate change and has backed away from Canada’s Kyoto commitment. In January, it muzzled Environment Canada’s scientists, ordering them to route all media enquires through Ottawa to control the agency’s media message. Last week, the prime minister and members of the cabinet failed to attend a ceremony to honour the Canadian scientists who contributed to the international climate-change report that won a share of the 2007 Nobel Peace Prize.
…
On the surface, funding for university-based research seems strong. The annual budgets for the Canadian Institutes of Health Research (CIHR) and the National Sciences and Engineering Research Council tripled and doubled, respectively, between 2000 and 2005. The government has also supported new science projects through government-created corporations such as Genome Canada and the Canada Foundation for Innovation, and has recruited and retained promising young scientists through the Canada Research Chairs programme.
But Genome Canada funds only half of the cost of a research project — scientists must seek the remaining cash from elsewhere. Last year, the CIHR was able to fund only 16% of the applications it received, and cut the budgets of successful applicants by a quarter, on average. And earlier this month, the country’s top scientists and university officials warned that they were short of funds to operate multimillion-dollar big-science projects such as the Canadian Light Source synchrotron.
What’s to be done? Canada has made good investments in its science infrastructure and its future research leaders. The present government might be dissolved after a vote of confidence next month, which could in itself lead to a change for the better. But in any circumstances, Canada’s leading scientists can be public advocates, pointing to the examples of other countries in urging the government of the day to boost their country into a position of leadership rather than reluctant follower.
Let me say this. I admit that I have been a beneficiary of some of the elitist components of the system, including graduate scholarships, a postdoctoral fellowship, and individual awards. I also have co-authored successful Genome Canada and Canada Foundation for Innovation grants. But this new focus on supporting few to the detriment of many is as un-Canadian as it is scientifically disastrous. It is my sincere hope that Canada’s researchers will refuse to accept this assault on science in our country.
Conservative government versus basic research.
First they dismissed the National Science Advisor and closed the office. Now they announce increases in funding for science — to a small elite, and only for those working in areas the Conservative government thinks are important. It’s all applied topics — for the agency that presently funds most basic science, NSERC: automotive, manufacturing, forestry, and fishing industries. Instead of raising support for everyone, which is much needed, they will give it to a small few. This is so… un-Canadian. One of the craziest bits is offering $50,000 per year to a select group of doctoral students — they could provide scholarships for twice as many at half the price per, and they would still be doing well compared to most students.
I hate that I had to start a new label, “politics”, just for this post, but I can barely take it.
From ScienceNow‘s report:
- “Since assuming office in 2006, Canada’s minority Conservative government has argued that it’s more important to fund the best and the brightest in designated areas than to spread the wealth across the entire spectrum of scientific activity. Today, it reinforced that message in a new 2008-2009 budget that will shower 20 scientific superstars from within Canada and abroad with $10 million apiece over 7 years.”
- “It has yet to be determined whether the chairs will be selected through competitions administered by the country’s three research granting councils or whether a government department such as Industry Canada will oversee the program, including selection of the recipients.”
- “Finance officials stressed that the allocations must be directed at priority areas rather than simply pumped into core operating grant programs. ‘The government has made it clear that they want this money to support the kind of research that should be supported,” says one official. ‘And the councils will have to answer to the Treasury Board if they don’t.'”
- “Doctoral students will also be beneficiaries of the move toward more elitism. Scholarships named in honor of war hero and former Governor General Georges P. Vanier will be created to attract 500 of ‘the best doctoral students from here and around the world to study in Canada’ each year. Each student will be eligible for $50,000 per year for up to 3 years.”
Expect a significant discussion among Canadian researchers — perhaps not even totally polite — over this.
Teaching evolution.
ERV points us to an interview in Wired where Carl Woese argues that we should stop teaching evolution in high school because it cannot be dealt with properly at this level. ERV is, um, unhappy with this. My take is posted here.
Quotes of interest — satellite DNA in the news.
I have already made note of some of the coverage of noncoding DNA that appeared in Science during the 1980s, and as a sequel to that earlier installment of the series, I want to talk about the coverage in Nature from the late 1960s and early 1970s. Because SINEs, LINEs, pseudogenes, and introns were all discovered in 1977 or later, this will necessarily focus on satellite DNAs.
As mentioned previously, satellite DNAs were discovered in the early 1960s, and by the late 1960s and early 1970s there was substantial interest in these highly repetitive components of the genome. Nature published several stories about this work in their “News and Views” section, authored by various unnamed correspondents. Of course, one must not take the interpretation of anonymous science writers as definitive (after all, their contemporary counterparts do much to add the the mythology surrounding noncoding DNA), but it supports the overall contention that during this period adaptationist thinking was dominant and thus that it was taken almost as a given that functions would be elucidated for noncoding sequences. You will notice also that many of these stories report on data that contradict proposed functions, yet the expectation remained that some function exists. I am not criticizing the studies of these early authors in any way. Some satellite DNA is functional in chromosomal structure, but the point is that at the time this was an a priori assumption rather than a conclusion, and it is clearly not the case that these elements were dismissed as unimportant.
“Mouse satellite DNA”, Nature 215: 575, August 5, 1967:
What is the function of satellite DNA? It is unlikely to code for protein and yet it forms 10 per cent of the cell’s total DNA. What possible purpose is served by having so many, apparently identical, short sequences within the same genome?
“Satellite DNA”, Nature 222: 327, April 26, 1969:
Unfortunately, the group’s latest data serve only to make ideas about the function of this strange DNA fraction even more obscure.
…
But if satellite DNA is not transcribed, what is its function? Flamm et al. are impressed by the fact that numerous copies of a nucleotide sequence of 350 bases have been maintained during evolution in the face of the tendency to accumulate random mutations. This implies that satellite DNA has some important function. They suggest that it is required for “housekeeping”, the folding and packing of DNA in the chromosomes. In the absence of any critical data or any way of testing for the function of this DNA, that is as good a suggestion as any.
…
Like the Edinburgh group, Maio and Schildkraut are convinced that satellite DNA has some vital, albeit unknown, function.
“Hybridization and satellite DNA”, Nature 225: 414, January 31, 1970:
The function of this satellite DNA has always been obscure, reducing investigators to suggest, for example, that it may be involved in chromosome “housekeeping”, but Pardue and Gall claim that it is localized in the centromeres. It may therefore play a role in chromosome pairing, and this may account for the curious properties of satellite DNA, not least its peculiar base sequence.
“Mysterious satellites”, Nature 225: 899-900, March 7, 1970:
Any biologist told that 10 to 12 percent of the total DNA genome of an animal is sequestered in a chemically distinct fraction would find it hard to escape the conclusion that such DNA has some crucial cellular function. That explains why the so-called satellite DNAs are exciting so much interest…
…
A host of experiments and speculations leap to mind. Perhaps satellite DNA plays some part in the assembly of the mitotic spindle, for example, by influencing polymerization of spindle protein or the attachment of chromosomes to the spindle. Hybrid cells might be useful in studying the specificity of a putative interaction between satellite DNA and components of the mitotic spindle. And the chromosomes of organisms with diffuse centromeres might be useful for further testing the relationship between satellite DNA and centromeres.
The last one is interesting, because it led to a correction by one of the first people to identify satellite DNA, Waclaw Szybalski, in the correspondence segment of the April 4, 1970 issue. Did he complain about the characterization of biologists anticipating functions for satellite DNA? No, he simply noted that the author got the date of discovery wrong (Szybalski 1970; Nature 226: 89-90).
“Satellite DNA and sequence”, Nature 227: 775, August 22, 1970:
What possible function can be served by a DNA which consists of tandem duplication of a sequence of only six base pairs, and why should an animal such as the guinea-pig require some 107 copies of this short sequence in all its cells?
…
Finally, even though we now know the basic sequence unit of a satellite DNA we are no closer to explaining the function of these specialized DNAs. Since they have no role in coding protein, the most plausible suggestion is that they have some role in maintaining the integrity of the chromosome itself. The localization of satellite DNA in the centromere regions of chromosomes suggests they play a part in the functions conventionally ascribed to the centromere. But for the time being such suggestions remain speculative.
“Satellite DNA and speciation”, Nature 240: 128, November 17, 1972:
The function and evolutionary significance of satellite DNA — DNA which has a reiterated base sequence, is associated with heterochromatin and centromeres and may or may not be transcribed — remain tantalizing mysteries. It seems unlikely that these simple sequences code for any polypeptides and it has, therefore, been suggested that satellite DNA may be involved in processes such as pairing of homologous chromosomes, chromosome movement and chromosome packing, but there is little evidence in support of these speculations.
“The mystery deepens”, Nature 240: 255, December 1, 1972:
But the fact remains that one is still at a loss as to the function of satellite DNA, the chief characteristic of which is its comparatively simple and highly reiterated base sequence, and indeed the more that is learnt about the distribution of satellite DNAs the deeper the enigma of its function becomes.
“DNA dominant at Berkeley, California”, Nature 245: 183-184, September 28, 1973:
The problem of DNA redundancy continues to intrigue several teams, without finally yielding all the secrets of its function or the reason for the wide variation in amount from species to species. Some of the extra DNA is almost certainly present as spacer sequences between cistrons, but this does not account for the large amount of simple sequence DNA, present in millions of copies, in the centromeric heterochromatin. P.M.B. Walker, for whom the Medical Research Council has recently set up a unit in Edinburgh specifically devoted to research on the mammalian genome, reviewed the history of satellite DNA, but said that most investigators would still go no further than suggest that this material, which is not transcribed, has some “housekeeping” function.
Here is the take-home message. From the time it was discovered, satellite DNA was presumed to be functional on the basis of Darwinian adaptationist expectations. This stimulated intensive research on the subject which was considered interesting enough to be reported about regularly in Nature. Some of the proposed functions, such as a structural role for some noncoding sequences, turned out to be correct — which was only shown because the Darwinian assumption prompted researchers to test functional ideas. The claim by creationists that “Darwinism” prevented such research is manifestly and demonstrably inaccurate. The problem, as I have noted, is that a strict focus on adaptive roles for noncoding DNA prevented many researchers from adopting a more balanced approach under which some of it is functional but most of it is not.
____________
Part of the Quotes of interest series.
Quotes of interest — SINEs and LINEs.
I am hopeful that our exploration of the peer-reviewed scientific literature and related news stories in scientific journals from the 1960s to the 1990s convincingly reveals that those who claim that junk DNA was “long dismissed as irrelevant” have it exactly backwards. Throughout this period, but especially before the non-adaptationist (though not exclusive) alternative offered by the selfish DNA hypothesis began to influence thinking on the topic by the mid-1980s, it was assumed, following Darwinian logic, that the very existence of so much DNA meant that it must be functional for the organism. It is only after considerable empirical investigation of potential functions that it became a common view that most (but certainly not all) noncoding DNA is unlikely to be functional at the level of the organismal phenotype.
I have already mentioned Alu elements — by far the most common single type of noncoding DNA element in the human genome. Alu elements are part of the category of repetitive DNA known as SINEs, which stands for short interspersed repeated sequences (or short interspersed nuclear elements). These sequences are now recognized as a type of transposable element that uses an RNA intermediate (i.e., undergoes retrotransposition) but which cannot do so without borrowing (some say parasitizing) the molecular transposition apparatus of other elements, namely long interspersed repeated sequences (LINEs). LINEs are not as common in the human genome as SINEs, but as they are much larger, they make up more of the total DNA. Whereas there are about 1.5 million SINEs (1 million of them Alu) making up about 13% of the genome sequence, the 870,000 or so copies of LINE elements (more than 500,000 of them LINE-1) constitute more than 20% of human DNA.
The terms SINE and LINE were coined by Maxine Singer in 1982 (Singer 1982a). By that time, the term “junk DNA” (Ohno 1972; Comings 1972) had been in circulation for a decade, and this was also two years after the “selfish DNA” hypothesis was put forward by Orgel and Crick (1980) and Doolittle and Sapienza (1980). Singer (1982b) cited these latter papers (but not Ohno’s) in her longer review of mammalian repeated DNA sequences. So once again, we have a prime candidate for assessing the general attitude in the scientific community regarding possible function of noncoding DNA sequences during the supposed period of neglect.
Were SINEs and LINEs dismissed as mere junk unworthy of further exploration?
Singer (1982a):
Function?
The critical question about SINEs and LINEs concerns their function, if they have any. The catalog of proposed functions for SINEs includes many of the unsolved problems in molecular biology, but none has been demonstrated directly. The existence of RNA transcripts from some SINE-family members is the most compelling argument available that they have a function, although functions independent of transcription (and in addition to transposition) have also been suggested. (The possibility that LINEs are transcribed requires investigation). Particularly striking is the fact that the 4.5S transcripts of Alu-like SINEs of hamster and mice are more than 95% identical in sequence, which is significantly closer than the variation among the different copies of a SINE family in a single species. If we assume that one or a few SINEs encode the 4.5S RNAs, is there any functional significance to the many other dispersed copies of family members? It seems reasonable to expect that there is some trade-off between an advantage imparted to cells by SINEs and the disadvantage of a promiscuous and abundant mobile element that is presumably destructive if implanted in an essential coding region.
Singer (1982b):
[A number of in-line citations have been omitted for clarity]Are SINES functional?
As a background, it is interesting to recall proposals suggesting that highly repeated dispersed sequences may be without function (Orgel and Crick 1980; Doolittle and Sapienza 1980) and also disagreement concerning those proposals (Cavalier-Smith 1980; Dover 1980; T.F. Smith 1980; Orgel et al. 1980; Dover and Doolittle 1980). Specific functions that have been suggested include the control of gene expression, perhaps by involvement of transcripts of SINES in the maturation of messenger RNA, and service as origins of DNA replication.…
The following additional point may be important, in view of the suggestions that highly repeated sequences have no function at all. A mobile element may generate diversity with a potential selective advantage, but it can also generate disadvantage if it moves into an essential gene. Mutation by movable elements has been demonstrated in yeast and Drosophila. The high frequency of mutation caused by the presence of large numbers of movable elements within a mammalian genome might have proven intolerable and been selected against, unless it was counterbalanced by some positive functional advantage.
Finally, the suggestion that SINES may serve as origins for DNA replication should be considered. The basis for the suggestion is the presence in SINES of a short (14bp) homology to a sequence associated with the origin of replication of murine and primate popaviruses. Georgiev et al. (1981) describe some preliminary experiments that are consistent with this suggestion. However, in popavirus genomes this region is part of a complex control region and may be involved in the control of transcription as well as replication. Only additional experiments will resolve these questions.
…
Are LINES functional?
The discovery of LINE families in mammals is recent and there is very little information available regarding function. Adams et al. (1980) found no transcripts homologous to the human Kpn-LINE family in bone marrow cells and Manuelidis [1982] also reports negative preliminary experiments. There is no information available regarding the possibility that LINES are mobile in mammalian genomes.
As noted previously, the SINE Alu was first described in 1979, and the first LINEs were discovered using similar methods around 1980. Singer (1982b) cites several publications and articles in press detailing sequences of this type from the human and mouse genomes. Most of these papers did not include any discussion one way or the other about function and focused instead on the technique used or the specific molecular characteristics of the sequences. However, one of the early papers did discuss function (and non-function).
Adams et al. (1980):
As to the function or genesis of this sequence we can make only vague hypotheses. The fact that it is not expressed into RNA, at least in bone marrow cells, at levels proportionate to its reiteration frequency, suggests that it does not code for a protein or major nuclear RNA in this tissue. However, there may be a low-level transcript which has some functional role, or there may be transcription in some other tissue. Alternatively this sequence may be a binding site for a chromosomal protein, or serve as a signal for chromosomal folding. As such it could conceivably have some role in the regulation of expression of the β-globin or other nearby genes. The interspersion of this sequence among other DNA is consistant with but not by itself supportive of such a role. Finally it is possible that this repeated sequence has no function relevant to the organism, but is carried in the genome in an essentially parasitic fashion (Doolittle and Sapienza 1980).
____________
Part of the Quotes of interest series.
____________
References cited
Adams, J.W., R.E. Kaufman, P.J. Kretschmer, M. Harrison, and A.W. Nienhuis. 1980. A family of long reiterated DNA sequences, one copy of which is next to the human beta globin gene. Nucleic Acids Research 8: 6113-6128.
Cavalier-Smith, T. 1980. How selfish is DNA? Nature 285: 617-618.
Comings, D.E. 1972. The structure and function of chromatin. Advances in Human Genetics 3: 237-431.
Doolittle, W.F. and C. Sapienza. 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284: 601-603.
Dover, G. and W.F. Doolittle. 1980. Modes of genome evolution. Nature 288: 646-647.
Georgiev, G.P., Y.V. Ilyin, V.G. Chmeliauskaite, A.P. Ryskov, D.A. Kramerov, K. G. Skryabin, A. S. Krayev, E. M. Lukanidin, and M. S. Grigoryan. 1981. Mobile dispersed genetic elements and other middle repetitive DNA sequences in the genomes of Drosophila and mouse: transcription and biological significance. Cold Spring Harbor Symposia on Quantitative Biology 45: 641-654.
Manuelidis, L. 1982. Repeated DNA sequences and nuclear structure. In Genome Evolution (eds. G. Dover and A. Flavell), pp. 263-285. Academic Press, New York.
Ohno, S. 1972. So much “junk” DNA in our genome. In Evolution of Genetic Systems (ed. H.H. Smith), pp. 366-370. Gordon and Breach, New York.
Orgel, L.E. and F.H.C. Crick. 1980. Selfish DNA: the ultimate parasite. Nature 284: 604-607.
Orgel, L.E., F.H.C. Crick, and C. Sapienza. 1980. Selfish DNA. Nature 288: 645-646.
Singer, M.F. 1982a. SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes. Cell 28: 433-434.
Singer, M.F. 1982b. Highly repeated sequences in mammalian genomes. International Review of Cytology 76: 67-112.
Smith, T.F. 1980. Occam’s razor. Nature 285: 620.
Someone thinks Genomicron is excellent!
I am always glad when people let me know that they appreciate the content of the blog. My posts tend not to generate much in the way of comments, and aside from the occasional email I don’t really have a good sense of how the blog is perceived. But every now and then a fellow blogger is kind enough to let me know in some especially interesting way. Case in point, the Golden Mouse Award that was kindly bestowed upon Genomicron by Bertalan Meskó of ScienceRoll.
Now, Ian Ramjohn of Further Thoughts has been kind enough to pass along the latest E for Excellence award. Thanks, Ian! Update: Adrian Thysse of Mystery of Mysteries has named Genomicron as well.

In return, I will point out that I have some of the brightest readers around.


