I have argued that prior to 1980, when the selfish DNA hypothesis was proposed, it was taken more or less as a given by most biologists that noncoding DNA had some function(s), even if the specific adaptive significance of these sequences had yet to be demonstrated. This was based on simple Darwinian, adaptationist logic: if it’s there, it must be doing something good for the organism or else natural selection would have done away with it a long time ago.
The point of the selfish DNA hypothesis was to provide an alternative view that did not preclude the potential function of some noncoding DNA, but also did not require it all to be adaptive for its presence to be accounted for. In reality, this simply shifted the process of natural selection down one level, from organisms in populations to sequences of DNA within the genome, but the consequence was important because it meant that evidence had to be provided for organism-level function — this could not just be assumed simply because noncoding DNA exists.
I have already quoted from many papers published during the 1980s, which showed that many biologists continued to explore possible functions for noncoding DNA of different kinds during the 1980s, as they continue to do today. In this post, I want to back up the claim that a common assumption during the 1970s was that most DNA in the genome is functional for the organism. By the late 1970s we start to see a shift away from this position and toward (untenable) ideas regarding “evolutionary” functions — but that is a topic for another time. Here, then, are some samples of the peer-reviewed literature from the period from 1970 to 1979.
At present, one can only speculate on the functions of much of the repeated DNA in eukaryotes. For example, repetitious DNA might contain binding sites for chromosomal proteins or chromosomal RNA… A certain portion of the repetitious DNA may play a role in determining how DNA fibers fold in the chromosome… A fraction of repetitious DNA may not contain any genetic “information” at all but may have evolved to function merely as space-filler in the genome.
[Note: this quote is heavily abbreviated from more than one paragraph, so please consult the original for the full text].
…
We can summarize these speculations about the genetic functions of repetitious DNA by concluding that, whatever they are, they are probably diverse. Regardless of the function of repetitious DNA, however, its very existence raises fundamental problems for [adaptationist] evolutionary theory.
Edelman, G.M. and J.A. Gally. 1970. Arrangement and evolution of eukaryotic genes. In The Neurosciences Second Study Program (ed. F.O. Schmitt), pp. 962-972. Rockefeller University Press, New York.
It was first demonstrated by Boivin and the Vendrelys that mammalian cells from different tissues contain the same amount of DNA, and furthermore that sperm cells contain half this amount. In several species of fungi vegetative diploid cells contain twice the amount of DNA contained by haploid ones. It is also well known that DNA is metabolically stable. These observations strongly suggest that most, if not all, the DNA has a genetic function. The difficulty arises when the actual amounts of DNA per nucleus in different species are considered.
…
The problem of large amounts of DNA per genome might be explained if only a proportion of the DNA had a genetic function and the rest was redundant or played some other role in the cell.
…
In summary, there appear to be several possible functions for DNA which does not encode for messenger RNA and protein synthesis. Direct information about such functions is negligible and one can only guess at the total amount of DNA which may be involved.
Holliday, R. 1970. The organization of DNA in eukaryotic chromosomes. Symposia of the Society for General Microbiology 20: 359-379.
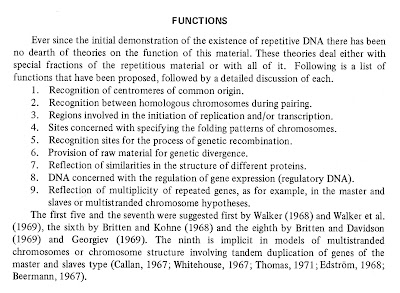
Alternative hypotheses concerning the function of repetitious DNA have recently been proposed, and it seems an appropriate time to review the evidence for the existence of repetitious DNA and see how it adds to our understanding of the evolution and organization of DNA within organisms.
 …
…
None of the recognition functions, i.e., recognition of centromeres, initiation sites, pairing sites, recombination sites, folding sites, or regulatory sites, that we have discussed is mutually exclusive of the others. They all relate to cellular phenomena that have been demonstrated or inferred from other data. All these phenomena probably exist within every higher organism. Therefore, DNA involved in each of these functions could contribute in varying degrees to the repeated portion of the genome.
Bostock, C. 1971. Repetitious DNA. Advances in Cell Biology 2: 153-223.
There is no doubt, however, that the presence of satellite DNA confers some selective advantage; otherwise we cannot account for its spread throughout a population as large as that of the house mouse.

The origin and spread through the population of such diverse sequences therefore present considerable problems, and I would like to suggest that the best explanation might be that satellite DNA confers a direct advantage on the chromosome which carries it, because such a chromosome survives the mechanical processes of, primarily, meiosis better than a sister chromosome not so well endowed.
Walker, P.M.B. 1971. Origin of satellite DNA. Nature 229: 306-308.
With the assumption that a portion that comprises some 10 percent of the genomes in higher organisms cannot be without a raison d’être, an extensive review led us to conclude that a certain amount of constitutive heterochromatin is essential in multicellular organisms at two levels of organization, chromosomal and nuclear. At the chromosomal level, constitutive heterochromatin is present around vital areas within the chromosomes. Around the centromeres, for example, heterochromatin is believed to confer protection and strength to the centromeric chromatin. Around secondary constrictions, heterochromatic blocks may ensure against evolutionary change of ribosomal cistrons by decreasing the frequency of crossing-over in these cistrons in meiosis and absorbing the effects of mutagenic agents. During meiosis heterochromatin may aid in the initial alignment of chromosomes prior to synapsis and may facilitate speciation by allowing chromosomal rearrangement and providing, through the species specificity of its DNA, barriers against cross-fertilization.
At the nuclear level of organization, constitutive heterochromatin may help maintain the proper spatial relationships necessary for the efficient operation of the cell through the stages of mitosis and meiosis. In the unicellular procaryotes, the presence of a small amount of genetic information in one chromosome obviates the need for constitutive heterochromatin and a nuclear membrane. At higher levels of organization, with an increase in the size of the genome and with evolution of cellular and sexual differentiation, the need for compartmentalization and structural components in the nucleus became imminent. The portion of the genome that was concerned with synthesis of ribosomal RNA was enlarged and localized in specific chromosomes, and the centromere became part of each chromosome when the mitotic spindle was developed in evolution. Concomitant with these changes in the genome, repetitive sequences in the form of constitutive heterochromatin appeared, probably as a result of large-scale duplication. The repetitive DNA’s were kept through natural selection because of their importance in preserving these vital regions and in maintaining the structural and functional integrity of the nucleus.
The association of satellite (or highly repetitive) DNA with constitutive heterochromatin is understandable, since it stresses the importance of the structural rather than transcriptional roles of these entities. Nuclear satellite DNA’s have one property in common despite their species specificity, namely heterochromatization. In this sense the apparent species specificity of satellite DNA may be the result of natural selection for duplicated short polynucleotide segments that are nontranscriptional and can be utilized in specific structural roles.
Yunis, J.J. and W.G. Yasmineh. 1971. Heterochromatin, satellite DNA, and cell function. Science 174: 1200-1209.
The great variability between species (and even populations) in the type and quantity of satellite and/or cryptic sequence DNA suggests that organisms function within wide limits for organizing the type and extent of constitutive heterochromatin. On the other hand, the striking species specificity of major satellite sequences, together with a rather low level of base sequence divergence within a species and the precise localization of much of this DNA to centric heterochromatin, suggest that these DNA’s perform important nuclear functions.
Rae, P.M.M. 1972. The distribution of repetitive DNA sequences in chromosomes. Advances in Cell and Molecular Biology 2: 109-149.
Today there are over 100 laboratories studying different aspects of the problem concerning highly repetitive sequences and, as is always the case, a new and often confusing jargon has grown up around the rapidly evolving field. It is the purpose of this chapter to define and describe these new terms and to focus on the following three questions: What are highly repetitive sequences? How did they evolve? What is their function?
Flamm, W.G. 1972. Highly repetitive sequences of DNA in chromosomes. International Review of Cytology 32: 1-51.
[Satellite DNAs in mice] are widely established in populations throughout the world and thus clearly cannot be relegated to a random and functionless quirk of evolution. Simple-sequence DNAs must obviously possess a certain role in the genome.
…
There is one major hope for making sense of the fact that many higher organisms seem to carry in every nucleus a large portion of their DNA that looks, superficially, to be completely worthless. This lies in the comparative approach. When do simple-sequence DNAs arise in evolution? Can we find two closely related species one with and one without a major block of heterochromatin?
Swift, H. 1973. The organization of genetic material in eukaryotes: progress and prospects. Cold Spring Harbot Symposia on Quantitative Biology 38: 963-979.
Alternatively, large variations in genome size could readily be accommodated if a high proportion of the DNA were used for functions other than coding for proteins. A number of such functions have been proposed and incorporated into hypothetical structures for the eukaryotic genome.
…
The outstanding problem presented by eukaryotic DNA is that of finding a role for these large fractions not used in coding for proteins or cytoplasmic RNAs.
Southern, E. 1974. Eukaryotic DNA. In MTP International Review of Science, Biochemistry Series One, Volume 6, Biochemistry of Nucleic Acids (ed. K. Burton), pp. 101-139. University Park Press, Baltimore.
Genetic redundancy is common in eukaryotes and thus must confer substantial selective advantages upon this group of organisms. One function of certain redundant DNA sequences (satellite DNAs) that are located in the centromeric heterochromatin may be to facilitate proper chromosome pairing and segregation.
Tartof, K.D. 1975. Redundant genes. Annual Review of Genetics 9: 355-385.
Proposed functions for satellite DNA were evaluated and formally set forth by Walker (1971) and have since been expanded by Mazrimas and Hatch (1972), Lagowski et al. (1973), Lee (1975), Bostock (1971), Walker (1972), and Comings (1972). In a masterful summary and evaluation of current ideas relating repeated DNA to the organization of the eukaryote chromosome (Cold Spring Harbour Symposia on Quantitative Biology 1973) Swift stated that the function of simple sequence DNAs not only appeared to have most investigators mystified, but that the present theories concerning their function were not accepted with much enthusiasm. He did, however, point out that “There is one major hope for making sense of the fact that many higher organisms seem to carry in every nucleus a large portion of their DNA that looks superficially to be completely worthless. This lies in the comparative approach. When do simple sequence DNAs arise in evolution? Can we find two closely related species one with and one without a major block of heterochromatin?”
Miklos, G.L.G. and R.N. Nankivell. 1976. Telomeric satellite DNA functions in regulating recombination. Chromosoma 56: 143-167.
Satellites constitute from 1% to 65% of the total DNA of numerous organisms, including that of animals, plants, and prokaryotes. Their existence has been known for about 15 years, but, although it is thought that they must be biologically important, with few exceptions … their functions are still largely in the realm of speculation. This remains true despite their ubiquity and, except for polytenized tissues, their constancy as a fraction of the total DNA in all tissues of the particular animal or plant species in which they are observed.
Skinner, D.M. 1977. Satellite DNA’s. BioScience 27: 790-796.
The most important aspect of satellite DNA remains the nature of its functions. Although a large body of data has been gathered concerning its structure, distribution and properties in several different organisms, most of these results have in fact neither supported nor disproved any one of the particular hypotheses of function (see Comings, 1972; Swift, 1973; Hsu, 1975; Miklos and Nankivell, 1976; for evaluations of functions). The most popular hypothesis on satellite DNA function has been, and still is, that satellite DNA is involved in some aspect of chromosome mechanics such as chromosome pairing.
Yamamoto, M. and G.L.G. Miklos. 1978. Genetic studies on heterochromatin in Drosophila melanogaster and their implications for the functions of satellite DNA. Chromosoma 66: 71-98.
In spite of the large amount of information which now exists on the structure of satellite DNA, it is clear that the central issue, namely, function, has not been directly tackled. Probably the most important reason for this unsatisfactory state of affairs has been the signal failure to approach the problem of function experimentally, despite the considerable effort that has gone toward elucidating structural properties. In part this refractory state of affairs stems from the assumption that a knowledge of function necessarily follows from a knowledge of structure. In part too it is explained by the fact that the properties of satellite DNA have been evaluated within the framework of prokaryotic dogma without sufficient consideration of the higher-order phenomena which characterize the biology of eukaryotes.
It appears very obvious that we have now reached a stage in satellite DNA research where additional structural analyses are not revealing the nature of its function — and indeed there is a very good reason for this. The initial success of the prokaryotic approach to genetic function was due to its manipulative aspects. This approach, involving perturbation of a system by mutation, deletion, substitution and translocation, proved critical. Only recently has a similar approach been applied specifically in investigating satellite DNA function, although an enormous literature exits on experimental and natural modifications of heterochromatin, which bear directly on this issue.
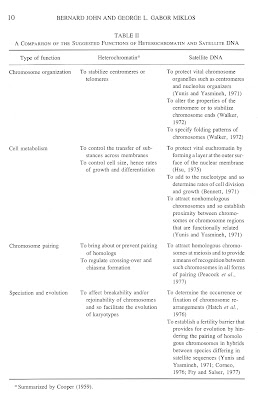
In the absence of experimental evidence the problem has in general been discussed in terms largely modified from earlier theoretical considerations relating to the functions of heterochromatin. A summary of the comparisons of heterochromatin and satellite DNA functions is presented in Table II. As can be seen from this table, the assumption has generally been made that there is at least one positive function. However, since similar organisms have widely different amounts of satellite DNA, and since such differences are found even between species that form viable hybrids, some investigators have suggested that these sequences are simply evolutionary by-products with no particular function. This fails to explain why so many eukaryotes have been found to contain highly repeated DNA and why its amount varies so considerably even between closely related species. Equally difficult to explain is why in some cases mechanisms have evolved to regulate replication of this DNA in particular tissues independently of the rest of the genome or indeed of other repetitious sequences in the same nucleus. Thus the large amount of satellite DNA in some species, its apparent rigid conservation in sequence and, as we shall see, its effects on the genome when it is altered in amount or position lead us to be unimpressed in general with the argument that most of it constitutes a functionless burden which many eukaryotes must bear. However, for the moment we will retain an open mind and examine the hard data pertaining to function before casting a final judgment at the end of this article.
 Click for larger image
Click for larger imageJohn, B. and G.L.G. Miklos. 1979. Functional aspects of satellite DNA and heterochromatin. International Review of Cytology 58: 1-114.
____________
Part of the Quotes of interest series.



