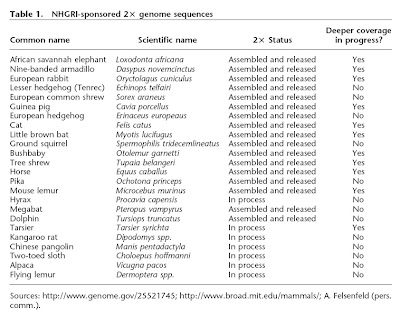
Lots of genomes going to be sequenced. Some of the members of the group are colleagues at Guelph. Very cool. That is all.
Genome 10K: A Proposal to Obtain Whole-Genome Sequence for 10 000 Vertebrate Species
Genome 10K Community of Scientists
The human genome project has been recently complemented by whole-genome assessment sequence of 32 mammals and 24 nonmammalian vertebrate species suitable for comparative genomic analyses. Here we anticipate a precipitous drop in costs and increase in sequencing efficiency, with concomitant development of improved annotation technology and, therefore, propose to create a collection of tissue and DNA specimens for 10 000 vertebrate species specifically designated for whole-genome sequencing in the very near future. For this purpose, we, the Genome 10K Community of Scientists (G10KCOS), will assemble and allocate a biospecimen collection of some 16 203 representative vertebrate species spanning evolutionary diversity across living mammals, birds, nonavian reptiles, amphibians, and fishes (ca. 60 000 living species). In this proposal, we present precise counts for these 16 203 individual species with specimens presently tagged and stipulated for DNA sequencing by the G10KCOS. DNA sequencing has ushered in a new era of investigation in the biological sciences, allowing us to embark for the first time on a truly comprehensive study of vertebrate evolution, the results of which will touch nearly every aspect of vertebrate biological enquiry.
http://www.nature.com/news/2009/091104/full/462021a.html